
RNA Editing Affects Repair of DNA Damage by the NEIL Enzyme
DNA Repair - Part 3
DNA damage is a phenomenon that can be detrimental to genomic integrity. Thankfully, our bodies have adapted many pathways to repair such DNA damage to prevent mutagenesis and cell death. There are many different topics related to DNA damage and repair, and I have recently focused on two other interesting topics related to this. In my first blog, I touched on the epigenetic role of DNA damage. In my second blog, I discussed ways to monitor DNA repair in a cellular context. To wrap up my three-part series, my last blog will focus on how RNA editing can affect repair of DNA damage by a unique family of DNA repair enzymes known as NEIL.
Adenosine deaminase acting on RNA 1 (ADAR1) is an enzyme capable of editing RNA after the RNA has been transcribed. It is capable of changing an adenosine base to an inosine base, which is read as guanosine during translation of the RNA (Figure 1).1 This changes the inherent code of the RNA, which can lead to changes in the final protein product. ADAR1 is known to edit a variety of different RNA transcripts, and this can lead to a variety of different consequences.

The NEIL family of enzymes is a DNA repair enzyme family that can remove a wide variety of DNA damage from a variety of different DNA contexts (Figure 2). The David Laboratory determined that the best characterized substrates for NEIL1 are the oxidative hydantoin lesions guanidinohydantoin (Gh) and spiriminodihydantoin (Sp). However, the NEIL enzymes are also capable of removing other modifications such as thymine glycol (Tg), 5-hydroxyuracil (5-OHU), and the formamidopyrimidine lesions (FapyG and FapyA). This is especially unique for an enzyme, as many enzymes are restricted to just one substrate. Additionally, many DNA repair enzymes are only capable of removing DNA damage from double-stranded DNA, but NEIL is unique in that it can repair damage from single-stranded DNA, bubble and bulge DNA (which might arise during replication and transcription), as well as G-quadruplex DNA. However, many of the initial studies of NEIL1 were conducted with only one isoform. Little did everyone know that there are actually two isoforms of NEIL1 that arise due to RNA editing!

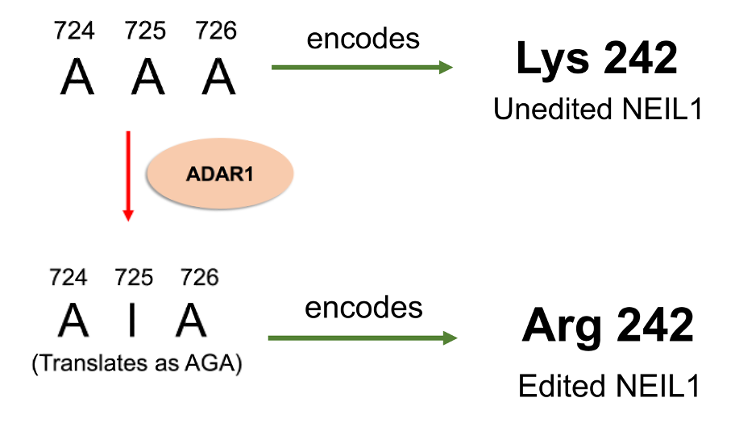
In 2009, a sequence analysis from human tissues was conducted to reveal possible adenosine to inosine editing sites by the ADAR protein. This was when the NEIL1 mRNA sequence was identified to contain an editing site. In 2010, the Beal and David laboratories then determined that position 725 of the NEIL1 mRNA can be edited by ADAR1. This codon event changes the AAA codon to AIA instead (Figure 3). Interestingly, this single codon change leads to a change in the amino acid in the protein. In the unedited (UE) protein, a lysine residue is placed at position 242, whereas in the edited (Ed) protein, an arginine is placed.
While you would think that this seemingly small change might not affect the way the enzyme processes DNA damage, position 242 of the protein is located at the beginning of the lesion recognition loop of the protein. Therefore, the Beal and David laboratories chose to analyze the ability of both isoforms (UE NEIL1 containing the lysine, and Ed NEIL1 containing the arginine) to repair a variety of different DNA damages. They used an in vitro glycosylase assay where DNA containing the damage is labelled with radioactive phosphorus. The DNA is then incubated with either Ed or UE NEIL1 and the reaction is quenched at various time points. Analysis of the base processing can be done with gel electrophoresis, and quantification yields the amount and rate of DNA repaired. The processing of Gh, Sp, and Tg was analyzed.
Interestingly, what was discovered is that Ed NEIL1 and UE NEIL1 process different DNA damages extremely differently! UE NEIL1 removes Tg about ~30-40-fold faster than Ed NEIL1 (Figure 4). In contrast, Ed NEIL1 removes Gh and Sp better, demonstrating ~3-4-fold greater activity. This is not due to a lack of binding, as binding results with non-cleavable Tg and Gh oligonucleotides shows similar affinities for both isoforms. The David laboratory then further examined a larger panel of oxidized purines and pyrimidines by both isoforms of NEIL1. From the analysis of these lesions, it was observed that UE NEIL1 prefers excising oxidized pyrimidines while Ed NEIL1 prefers oxidized purines.

Figure 4. Rates of base removal by edited (Ed) and unedited (UE) NEIL proteins. Tg, Gh, and Sp were analyzed via in vitro glycosylase assays and rates are listed above the bars. Ed NEIL1 is in blue and UE NEIL1 is in orange.
In 2016, the Chengqi laboratory published the first crystal structure of both Ed and UE NEIL1 bound to Tg-containing duplex DNA. What they discovered is that the lesion recognition loop actually undergoes a huge conformational change to contact the DNA damage. This demonstrated that the side chain of position 242, either lysine or arginine, directly interacts with the lesion. They proposed that tautomerization of Tg encouraged by amino acid contacts in the active site and protonation of N3 of the Tg base by either lysine or arginine enhances base excision. The lower pKa of lysine would make it a better proton donor during the cleavage step and is the proposed reason for the difference in excision of Tg by the two isoforms.
While the biological implication of having two isoforms of NEIL1 is not well understood, this may play an important regulatory role in DNA repair. ADAR1 editing produces two enzyme isoforms from a single gene with oxidized pyrimidines being excised much faster by UE NEIL1 and oxidized purines by Ed NEIL1. ADAR1 editing appears to maintain an important balance between the two isoforms in the cell. However, ADAR1 overexpression is associated with cancer. Under these conditions, the balance of Ed and UE NEIL1 could be shifted to express more of the edited isoform, leading to possible mutations, replication blocks, and strand breaks. Further investigation will be needed to reveal the impacts of having one isoform over the other. Clearly, the balance of Ed and UE NEIL1 within the cell plays a critical role in efficiently removing all DNA damage from the genome.
References
Anadon, C.; Guil, S.; Simo-Riudalbas, L.; Moutinho, C.; Setien, F.; Martinez-Cardus, A.; Moran, S.; Villanueva, A.; Calaf, M.; Vidal, A.; et al. Gene Amplification-Associated Overexpression of the RNA Editing Enzyme ADAR1 Enhances Human Lung Tumorigenesis. Oncogene 2016, 35 (33), 4407–4413. https://doi.org/10.1038/onc.2015.469.
Cao, S.; Rogers, J.; Yeo, J.; Anderson-Steele, B.; Ashby, J.; David, S. S. 2′-Fluorinated Hydantoins as Chemical Biology Tools for Base Excision Repair Glycosylases. ACS Chem. Biol. 2020, 15 (4), 915–924. https://doi.org/10.1021/acschembio.9b00923.
Hazra, T. K.; Izumi, T.; Boldogh, I.; Imhoff, B.; Kow, Y. W.; Jaruga, P.; Dizdaroglu, M.; Mitra, S.; Mutm, N. T. H. E. Identification and Characterization of a Human DNA Glycosylase for Repair of Modified Bases in Oxidatively Damaged DNA. Proc. Natl. Acad. Sci. 2002, 99 (6), 3523–3528. https://doi.org/10.1073/pnas.062053799.
Krishnamurthy, N.; Zhao, X.; Burrows, C. J.; David, S. S. Superior Removal of Hydantoin Lesions Relative to Other Oxidized Bases by the Human DNA Glycosylase HNEIL1. Biochemistry 2008, 47 (27), 7137–7146. https://doi.org/10.1021/bi800160s.
Li, J. B.; Levanon, E. Y.; Yoon, J.-K.; Aach, J.; Xie, B.; Leproust, E.; Zhang, K.; Gao, Y.; Church, G. M. Genome-Wide Identification of Human RNA Editing Sites by Parallel DNA Capturing and Sequencing. Science 2009, 324 (5931), 1210–1213. https://doi.org/10.1126/science.1170995.
Lotsof, E. R.; Krajewski, A. E.; Anderson-Steele, B.; Rogers, J.; Zhang, L.; Yeo, J.; Conlon, S. G.; Manlove, A. H.; Lee, J. K.; David, S. S. NEIL1 Recoding Due to RNA Editing Impacts Lesion-Specific Recognition and Excision. J. Am. Chem. Soc. 2022. https://doi.org/10.1021/JACS.2C03625.
Nishikura, K. A-to-I Editing of Coding and Non-Coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17 (2), 83–96. https://doi.org/10.1038/nrm.2015.4.
Teoh, P. J.; An, O.; Chung, T.-H.; Chooi, J. Y.; Toh, S. H. M.; Fan, S.; Wang, W.; Koh, B. T. H.; Fullwood, M. J.; Ooi, M. G.; et al. Aberrant Hyperediting of the Myeloma Transcriptome by ADAR1 Confers Oncogenicity and Is a Marker of Poor Prognosis. Blood 2018, 132 (12), 1304–1317. https://doi.org/10.1182/blood-2018-02-832576.
Yeo, J.; Goodman, R. A.; Schirle, N. T.; David, S. S.; Beal, P. A. RNA Editing Changes the Lesion Specificity for the DNA Repair Enzyme NEIL1. Proc Natl Acad Sci U S A 2010, 107 (48), 20715–20719. https://doi.org/10.1073/pnas.1009231107.
Yeo, J.; Lotsof, E. R.; Anderson-Steele, B. M.; David, S. S. RNA Editing of the Human DNA Glycosylase NEIL1 Alters Its Removal of 5-Hydroxyuracil Lesions in DNA. Biochemistry 2021, 28, acs.biochem.1c00062. https://doi.org/10.1021/acs.biochem.1c00062.
Zhu, C.; Lu, L.; Zhang, J.; Yue, Z.; Song, J.; Zong, S.; Liu, M.; Stovicek, O.; Gao, Y. Q.; Yi, C. Tautomerization-Dependent Recognition and Excision of Oxidation Damage in Base-Excision DNA Repair. Proc. Natl. Acad. Sci. 2016, 113 (28), 7792–7797. https://doi.org/10.1073/pnas.1604591113.
