How Does RNA Editing Affect Human Health?
How Does RNA Editing Affect Human Health?
by SeHee Park
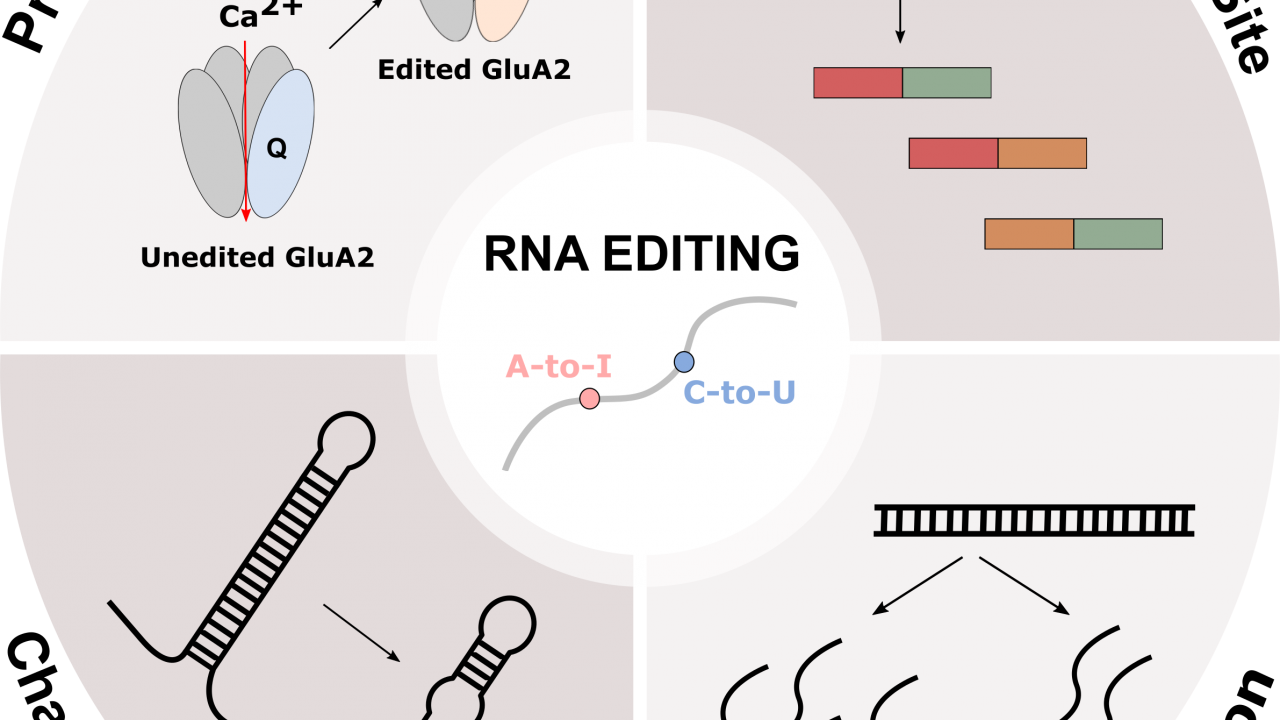
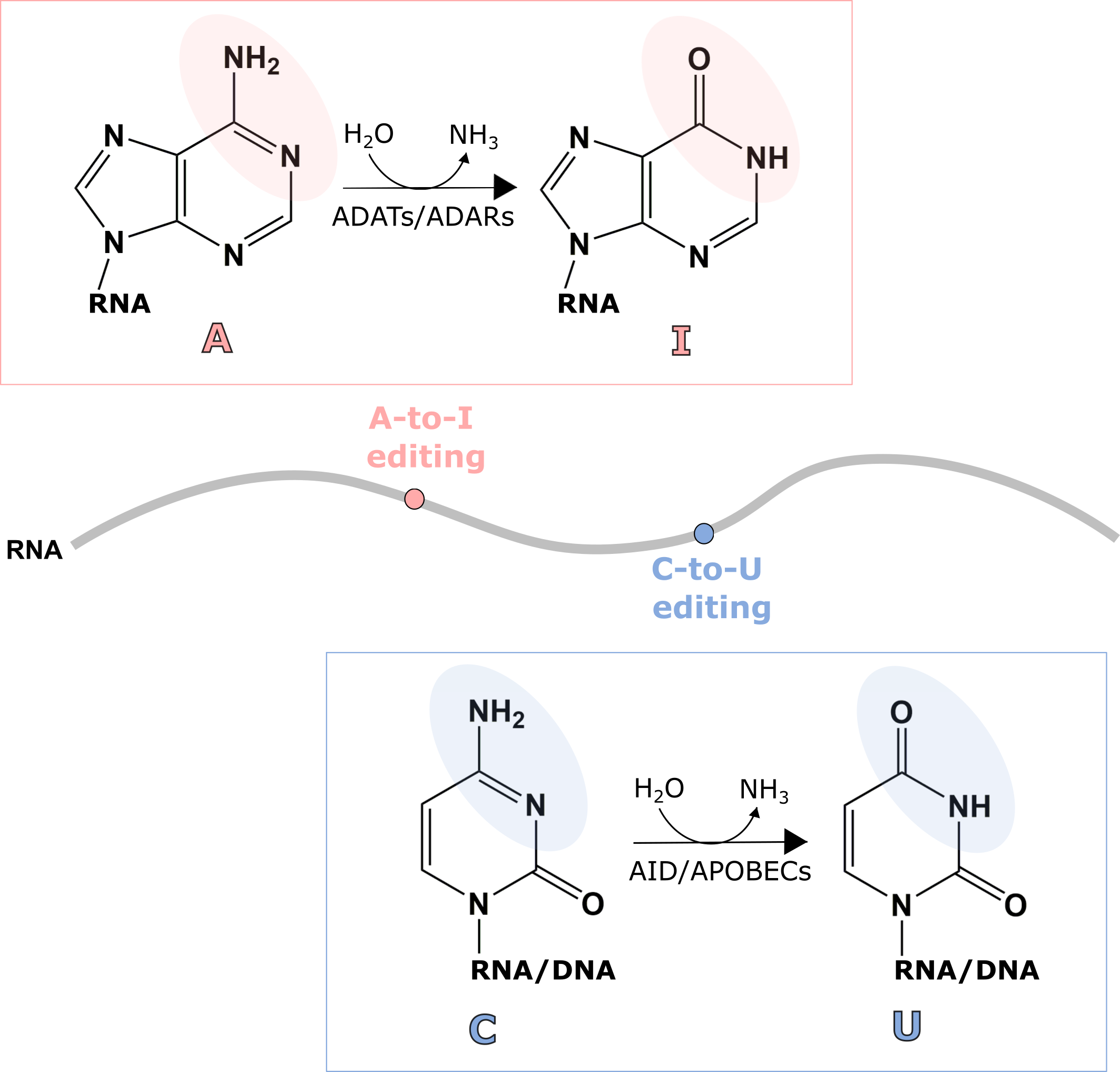
Ribonucleic acid (RNA) is one of the essential biomolecules that plays important roles in many cellular processes including protein synthesis, a process known as translation. RNA is made up of four types of nucleosides: Adenosine (A), Guanosine (G), Uridine (U), and Cytidine (C). In order to perform various biological functions, the nucleosides in RNA are often modified resulting in an expansion of nucleosides varying from the four classic RNA nucleosides: A, G, U, and C. RNA editing is the process that alters these components of RNA by insertion, deletion, or modification. There are two common types of RNA editing observed in humans: Adenosine to Inosine (A-to-I) modification and Cytidine to Uridine (C-to-U) modification. These modifications are catalyzed by deaminase proteins through hydrolytic deamination, which requires a water molecule.
Why should we care about these RNA modifications? We care because RNA is involved in various cellular processes and RNA modifications affect these processes. In other words, if these modifications are not regulated properly, it will affect its biological functions which in turn, could lead to various human diseases such as cancer.
C-to-U editing
In humans, AID (Activation-induced deaminase)/APOBEC (apolipoprotein B mRNA editing catalytic polypeptide-like) family of proteins are responsible for C-to-U modifications. APOBEC1 (apolipoprotein B mRNA editing enzyme catalytic subunit 1) was first identified by the discovery of C-to-U modification present in apolipoprotein B (apoB) mRNA. C-to-U substitution in apoB mRNA changes glutamate codon (CAA) to a stop codon (UAA), resulting in the expression of two different forms of protein that have a different biological property. After the discovery of APOBEC1, 10 other proteins that convert C-to-U were also discovered in humans: AID (Activation-induced deaminase), APOBEC2, APOBEC3A-H, and APOBEC4 proteins. Interestingly, APOBECs are not only responsible for C-to-U modification within RNA, but also deoxycytidine to deoxyuridine (dC-to-dU) within DNA. AID plays an essential role in antibody diversification and APOBECs activity is important for viral immune defense.
A-to-I editing
Unlike C-to-U editing, the deamination of adenosine (A) within RNA produces inosine (I), which is not a part of the four major components of RNA. Nevertheless, inosine is recognized as guanosine by cellular machinery because it has a similar base pairing ability as that of guanosine. In humans, A-to-I modification is catalyzed by two main families of enzymes, ADATs (Adenosine deaminase acting on tRNA) and ADARs (Adenosine deaminase acting on dsRNA). Each deaminase is responsible for A-to-I modification of different types of RNA. ADATs convert adenosine to inosine within tRNA. Because tRNA is directly involved in protein synthesis, this change in tRNA can affect protein synthesis. ADARs are responsible for A-to-I modification within mRNA or non-coding RNA. ADAR was first identified by the discovery of the unwinding of double-stranded RNA (dsRNA) in eggs of African clawed frog (Xenopus laevis). Later, it was found that this unwinding of dsRNAs was caused by A-to-I modification catalyzed by ADAR. There are three different types of ADARs in humans: ADAR1, ADAR2, and ADAR3. ADAR activity is important for proper nervous system functioning in humans and abnormal editing has been linked to neurological disorders as well as cancer development.
Consequences of RNA editing
RNA editing can affect various cellular processes in several ways because it changes the information contents within RNA. One of the major impacts of RNA editing is protein recoding. Recoding is the process in which one or more nucleotide changes in RNA results in a different codon. This produces proteins that are different from their genetic forms and these different forms of proteins often have a modified function or structure. For example, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors are glutamate ionotropic receptors that plays important role in the central nervous system (CNS). It is composed of four different subunits (GluA1, GluA2, GluA3, and GluA4) and when it is activated by glutamate, which is the major neurotransmitter in CNS, it allows the influx of Na+ or Ca2+ ions. GluA2 mRNA which encodes for one of the glutamate receptor subunits is edited by ADAR. This ADAR mediated editing replaces a glutamine (Q) codon (CAG) with an arginine (R) codon (CIG is read as CGG) which is why this editing site is referred to as the Q/R site. The resulting edited GluA2 is impermeable to Ca2+ ions because arginine is a large and positively charged amino acid. This protein recoding event mediated by ADAR is important because if GluA2 is not edited, it causes a high influx of Ca2+ ions which is toxic to cells. For this reason, GluA2 mRNA in humans is nearly 100% edited and unedited GluA2 mRNA has been linked to neurodegenerative diseases such as Amyotrophic Lateral Sclerosis (ALS).

RNA editing affects more than just recoding of proteins, but a wide array of other cellular processes. A-to-I or C-to-U modification can change RNA structure resulting in a change in the stability of RNA as well as interactions with RNA binding proteins. RNA splicing, a process that is required to generate mature mRNA, is also influenced by RNA editing. Furthermore, it can also affect the gene expression level through changes in RNA interference (RNAi) pathways.
Since RNA editing has an impact on various biological processes, there have been studies showing that aberrant activities of deaminase proteins can lead to various human diseases such as cancer and neurological disorders. In fact, higher levels of RNA editing and high expression of these proteins are often found in cancer. Therefore, modulating RNA editing mediated by these proteins can be a promising therapeutic strategy for cancer treatment.
References
Christofi, T.; Zaravinos, A. RNA Editing in the Forefront of Epitranscriptomics and Human Health. J. Transl. Med. 2019, 17 (1), 319. https://doi.org/10.1186/s12967-019-2071-4.
(Conticello, S. G. The AID/APOBEC Family of Nucleic Acid Mutators. Genome Biol. 2008, 9 (6), 229. https://doi.org/10.1186/gb-2008-9-6-229.
Keegan, L. P.; Gallo, A.; O’Connell, M. A. The Many Roles of an RNA Editor. Nat. Rev. Genet. 2001, 2 (11), 869–878. https://doi.org/10.1038/35098584.
Slotkin, W.; Nishikura, K. Adenosine-to-Inosine RNA Editing and Human Disease. Genome Med. 2013, 5 (11), 105. https://doi.org/10.1186/gm508.
